Laplacian
We are interested in this section in the conforming finite element approximation of the following problem:
| \(\partial \Omega_D\), \(\partial \Omega_N\) and \(\partial \Omega_R\) can be empty sets. In the case \(\partial \Omega_D =\partial \Omega_R = \emptyset\), then the solution is known up to a constant. |
| Hereafter, we note \(\Gamma_D=\partial \Omega_D\), \(\Gamma_N=\partial \Omega_N\) and \(\Gamma_R=\partial \Omega_R\). |
|
In the implementation presented later, \(\partial \Omega_D =\partial \Omega_N = \partial \Omega_R = \emptyset\), then we set Dirichlet boundary conditions all over the boundary. The problem then reads like a standard laplacian with inhomogeneous Dirichlet boundary conditions: |
1. Variational formulation
We assume that \(f, h, l \in L^2(\Omega)\). The weak formulation of the problem then reads:
2. Conforming Approximation
We now turn to the finite element approximation using Lagrange finite element. We assume \(\Omega\) to be a segment in 1D, a polygon in 2D or a polyhedron in 3D. We denote \(V_\delta \subset H^1(\Omega)\) an approximation space such that \(V_{g,\delta} \equiv P^k_{c,\delta}\cap H^1_{g,\Gamma_D}(\Omega)\).
The weak formulation reads:
| from now on, we omit \(\delta\) to lighten the notations. Be careful that it appears both the geometrical and approximation level. |
3. Feel++ Implementation
In Feel++, \(V_{g,\delta}\) is not built but rather \(P^k_{c,\delta}\).
| The Dirichlet boundary conditions can be treated using different techniques and we use from now on the elimination technique. |
We start with the mesh
auto mesh = loadMesh(_mesh=new Mesh<Simplex<FEELPP_DIM,1>>);
the keyword auto enables type inference, for more details see Wikipedia C++11 page.
|
auto Vh = Pch<2>( mesh ); (1)
auto u = Vh->element("u"); (2)
auto mu = expr(soption(_name="functions.mu")); // diffusion term (3)
auto f = expr( soption(_name="functions.f"), "f" ); (4)
auto r_1 = expr( soption(_name="functions.a"), "a" ); // Robin left hand side expression (5)
auto r_2 = expr( soption(_name="functions.b"), "b" ); // Robin right hand side expression (6)
auto n = expr( soption(_name="functions.c"), "c" ); // Neumann expression (7)
auto solution = expr( checker().solution(), "solution" ); (8)
auto g = checker().check()?solution:expr( soption(_name="functions.g"), "g" ); (9)
auto v = Vh->element( g, "g" ); (3)Next we define
| 1 | the discrete space Vh=Pch<k>(mesh) \(\equiv P^k_{c,h}\), |
| 2 | an element u of Vh |
| 3 | an expression mu that is used as the diffusion coefficient |
| 4 | an expression f that is used for the right hand-side term in the equation |
| 5 | an expression r_1 that is used for the first term in the Robin boundary condition |
| 6 | an expression r_2 that is used for the first term in the Robin boundary condition |
| 7 | an expression n that is used to define the Neumann boundary condition |
| 8 | an expression solution that is used to define the exact solution if there is one |
| 9 | an expression g that is used to define the Dirichlet boundary condition. |
|
at the following line
|
the variational formulation is implemented below, we define the
bilinear form a and linear form l and we set strongly the
Dirichlet boundary conditions with the keyword on using
elimination. If we don’t find Dirichlet, Neumann or Robin in the
list of physical markers in the mesh data structure then we impose
Dirichlet boundary conditions all over the boundary.
tic();
auto l = form1( _test=Vh );
l = integrate(_range=elements(mesh),
_expr=f*id(v));
l+=integrate(_range=markedfaces(mesh,"Robin"), _expr=r_2*id(v));
l+=integrate(_range=markedfaces(mesh,"Neumann"), _expr=n*id(v));
toc("l");
tic();
auto a = form2( _trial=Vh, _test=Vh);
tic();
a = integrate(_range=elements(mesh),
_expr=mu*inner(gradt(u),grad(v)) );
toc("a");
a+=integrate(_range=markedfaces(mesh,"Robin"), _expr=r_1*idt(u)*id(v));
a+=on(_range=markedfaces(mesh,"Dirichlet"), _rhs=l, _element=u, _expr=g );
//! if no markers Robin Neumann or Dirichlet are present in the mesh then
//! impose Dirichlet boundary conditions over the entire boundary
if ( !mesh->hasAnyMarker({"Robin", "Neumann","Dirichlet"}) )
a+=on(_range=boundaryfaces(mesh), _rhs=l, _element=u, _expr=g );
toc("a");|
We have the following correspondence:
More details can be found in Mathematical keywords of Feel++ and the Mesh reference. |
next we solve the algebraic problem
tic();
//! solve the linear system, find u s.t. a(u,v)=l(v) for all v
if ( !boption( "no-solve" ) )
a.solve(_rhs=l,_solution=u);
toc("a.solve");next we compute the \(L^2\) norm of \(u_\delta-g\), it could serve as an \(L^2\) error if \(g\) was manufactured to be the exact solution of the Laplacian problem.
// compute l2 and h1 norm of u-u_h where u=solution
auto norms = [=]( std::string const& solution ) ->std::map<std::string,double>
{
tic();
double l2 = normL2(_range=elements(mesh), _expr=idv(u)-expr(solution) );
toc("L2 error norm");
tic();
double h1 = normH1(_range=elements(mesh), _expr=idv(u)-expr(solution), _grad_expr=gradv(u)-grad<2>(expr(solution)) );
toc("H1 error norm");
return { { "L2", l2 }, { "H1", h1 } };
};
int status = checker().runOnce( norms, rate::hp( mesh->hMax(), Vh->fe()->order() ) );and finally we export the results, by default it is in the ensight gold format and the files can be read with Paraview and Ensight. We save both \(u\) and \(g\).
tic();
auto e = exporter( _mesh=mesh );
e->addRegions();
e->add( "uh", u );
if ( checker().check() )
{
v.on(_range=elements(mesh), _expr=solution );
e->add( "solution", v );
}
e->save();
toc("Exporter");The full listing is available below
feelpp_qs_laplacian_2d and feelpp_qs_laplacian_3d#include <feel/feel.hpp>
int main(int argc, char**argv )
{
using namespace Feel;
using Feel::cout;
po::options_description laplacianoptions( "Laplacian options" );
laplacianoptions.add_options()
( "no-solve", po::value<bool>()->default_value( false ), "No solve" )
;
Environment env( _argc=argc, _argv=argv,
_desc=laplacianoptions,
_about=about(_name="qs_laplacian",
_author="Feel++ Consortium",
_email="feelpp-devel@feelpp.org"));
tic();
auto mesh = loadMesh(_mesh=new Mesh<Simplex<FEELPP_DIM,1>>);
toc("loadMesh");
tic();
auto Vh = Pch<2>( mesh ); (1)
auto u = Vh->element("u"); (2)
auto mu = expr(soption(_name="functions.mu")); // diffusion term (3)
auto f = expr( soption(_name="functions.f"), "f" ); (4)
auto r_1 = expr( soption(_name="functions.a"), "a" ); // Robin left hand side expression (5)
auto r_2 = expr( soption(_name="functions.b"), "b" ); // Robin right hand side expression (6)
auto n = expr( soption(_name="functions.c"), "c" ); // Neumann expression (7)
auto solution = expr( checker().solution(), "solution" ); (8)
auto g = checker().check()?solution:expr( soption(_name="functions.g"), "g" ); (9)
auto v = Vh->element( g, "g" ); (3)
toc("Vh");
tic();
auto l = form1( _test=Vh );
l = integrate(_range=elements(mesh),
_expr=f*id(v));
l+=integrate(_range=markedfaces(mesh,"Robin"), _expr=r_2*id(v));
l+=integrate(_range=markedfaces(mesh,"Neumann"), _expr=n*id(v));
toc("l");
tic();
auto a = form2( _trial=Vh, _test=Vh);
tic();
a = integrate(_range=elements(mesh),
_expr=mu*inner(gradt(u),grad(v)) );
toc("a");
a+=integrate(_range=markedfaces(mesh,"Robin"), _expr=r_1*idt(u)*id(v));
a+=on(_range=markedfaces(mesh,"Dirichlet"), _rhs=l, _element=u, _expr=g );
//! if no markers Robin Neumann or Dirichlet are present in the mesh then
//! impose Dirichlet boundary conditions over the entire boundary
if ( !mesh->hasAnyMarker({"Robin", "Neumann","Dirichlet"}) )
a+=on(_range=boundaryfaces(mesh), _rhs=l, _element=u, _expr=g );
toc("a");
tic();
//! solve the linear system, find u s.t. a(u,v)=l(v) for all v
if ( !boption( "no-solve" ) )
a.solve(_rhs=l,_solution=u);
toc("a.solve");
tic();
auto e = exporter( _mesh=mesh );
e->addRegions();
e->add( "uh", u );
if ( checker().check() )
{
v.on(_range=elements(mesh), _expr=solution );
e->add( "solution", v );
}
e->save();
toc("Exporter");
// compute l2 and h1 norm of u-u_h where u=solution
auto norms = [=]( std::string const& solution ) ->std::map<std::string,double>
{
tic();
double l2 = normL2(_range=elements(mesh), _expr=idv(u)-expr(solution) );
toc("L2 error norm");
tic();
double h1 = normH1(_range=elements(mesh), _expr=idv(u)-expr(solution), _grad_expr=gradv(u)-grad<2>(expr(solution)) );
toc("H1 error norm");
return { { "L2", l2 }, { "H1", h1 } };
};
int status = checker().runOnce( norms, rate::hp( mesh->hMax(), Vh->fe()->order() ) );
// exit status = 0 means no error
return !status;
}4. Test Cases
We are now ready to test this Laplacian Feel++ Implementation with 2D and 3D test cases.
The test cases are available in Docker in the directory (/usr/local/share/feelpp/testcases/quickstart/laplacian).
|
4.1. Circle
Circle is a 2D testcase where \(\Omega \subset \RR^2\) is a disk whose boundary
has been split such that \(\partial \Omega=\partial \Omega_D \cup
\partial \Omega_N \cup \partial \Omega_R\).
In the following, we consider a manufactured solution \(u=x^2+y^2\) and we would like to verify the a priori convergence finite element property.
| This test case is tricky because the \(\partial \Omega\) is curved and \(\partial \Omega \neq \partial \Omega_h\). Depending on the boundary condition type and in particular the definition of the normal, we may obtain the proper convergence rate or not. |
We provide here the basic geometry used in Gmsh to describe \(\Omega\).
h=0.1;
Point(1) = {0, 0, 0, h};
Point(2) = {1, 0, 0, h};
Point(3) = {-1, 0, 0, h};
Point(4) = {0, 1, 0, h};
Point(5) = {0, -1, 0, h};
Circle(1) = {2, 1, 4};
Circle(2) = {4, 1, 3};
Circle(3) = {3, 1, 5};
Circle(5) = {5, 1, 2};
Line Loop(6) = {1, 2, 3, 5};
Plane Surface(7) = {6};
The Circle curve has several sections (1,2,3,5) that will be used to define the different boundary conditions, Dirichlet, Neumann and Robin.
|
4.1.1. The case \(\partial \Omega=\partial \Omega_D\)
We start with Dirichlet conditions on \(\partial \Omega\) which means that \(\partial \Omega_N = \partial \Omega_R = \emptyset\). To this end, we mark all boundary sections (1,2,3,5) as Dirichlet as shown in the Gmsh geo file below
circle-dirichlet.geoPhysical Line("Dirichlet") = {1, 2, 3, 5};
Physical Surface(8) = {7};The next step is to provide the proper conditions or expressions to Feel++ application feelpp_qs_laplacian_2d. They are described in the .cfg file.
circle-dirichlet.cfg[functions]
# Dirichlet
g=x^2+y^2:x:y
# right hand side
f=-4
# Robin left hand side
a=1
# Robin right hand side
b=2*(x*nx+y*ny)+x^2+y^2:x:y:nx:ny
# Neumann
c=2*(x*nx+y*ny):x:y:nx:ny
# mu: diffusion term (laplacian) (1)
mu=1| In the Dirichlet case, we do not have the issue mentioned above regarding the definition of the boundary and in particular the unit exterior normal to the boundary. We require only a point-wise evaluation of the condition on the boundary which are numerically exact. |
We are now ready to run the application.
$ cd /usr/local/share/feelpp/testcases/quickstart/laplacian/circle
$ mpirun -np 4 feelpp_qs_laplacian_2d --config-file circle-dirichlet.cfgThe execution should look like this:
Reading /usr/local/share/feelpp/testcases/quickstart/laplacian/circle/circle-dirichlet.cfg...
[ Starting Feel++ ] application qs_laplacian version 0.104.0-alpha.40 date 2018-Apr-17
. qs_laplacian files are stored in /feel/qs_laplacian/circle-dirichlet/np_1
.. logfiles :/feel/qs_laplacian/circle-dirichlet/np_1/logs
[loadMesh] Loading mesh in format geo+msh: "/usr/local/share/feelpp/testcases/quickstart/laplacian/circle/circle-dirichlet.geo"
[loadMesh] Use default geo desc: /usr/local/share/feelpp/testcases/quickstart/laplacian/circle/circle-dirichlet.geo 0.1
[loadMesh] Time : 0.0689365s
[Vh] Time : 0.0120969s
[l] Time : 0.0180983s
[a] Time : 0.0378079s
[a] Time : 0.0507547s
[a.solve] Time : 0.0267429s
[Exporter] Time : 0.0319186s
[L2 error norm] Time : 0.00716316s
[H1 error norm] Time : 0.0263121s
Checker exact verification failed for ||u-u_h||_H1
Computed error 1.39788e-14 (1)
Tolerance 1e-15
Checker exact verification failed for ||u-u_h||_L2
Computed error 1.54287e-15 (2)
Tolerance 1e-15
[env] Time : 0.246247s
[ Stopping Feel++ ] application qs_laplacian execution time 0.246247s| 1 | provides the \(H^1\) norm of the error with respect to the exact solution \(x^2+y^2\) |
| 2 | provides the \(L^2\) norm of the error with respect to the exact solution \(x^2+y^2\) |
| The error norms reached machine precision, the solution is in the finite element space. It is due to the fact that (i) we used \(P^2\) finite elements, (ii) the solution is itself quadratic and (iii) we used Dirichlet boundary conditions which requires only point-wise evaluation of the solution on the curved boundary. The fact that the boundary is curved do not affect the numerical solution. It won’t be the case in the presence of Neumann and Robin conditions. |
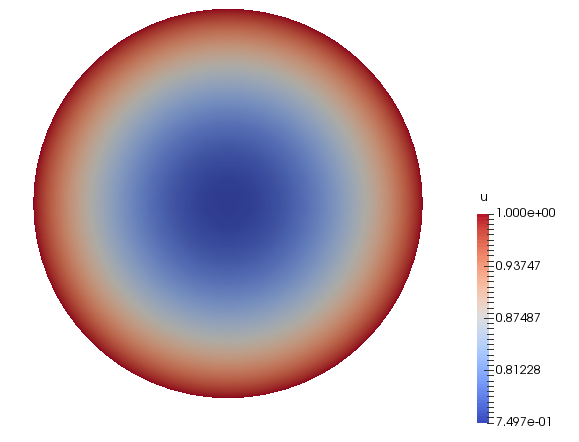
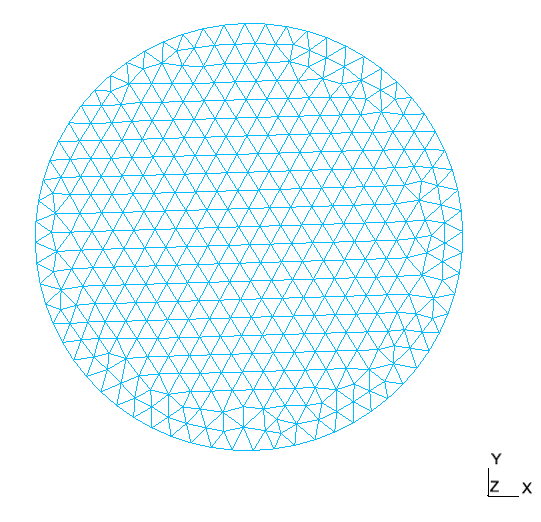
This gives us some data such as solution of our problem or the mesh used in the application which we can visualize using Paraview.
|
|
Solution \(u_\delta\) |
Mesh |
4.1.2. The case \(\partial \Omega=\partial \Omega_N\)
We continue with Neumann conditions on \(\partial \Omega\) which means that \(\partial \Omega_D = \partial \Omega_R = \emptyset\). To this end, we mark all boundary sections (1,2,3,5) as Neumann as shown in the Gmsh geo file below
circle-neumann.geoPhysical Line("Neumann") = {1, 2, 3, 5};
Physical Surface(8) = {7};The next step is to provide the proper conditions or expressions to Feel++ application feelpp_qs_laplacian_2d. They are described in the .cfg file.
circle-neumann.cfg[functions]
# dirichlet
g=x^2+y^2:x:y
# right hand side
f=-4
# robin
a=1
b=2*(x*nx+y*ny)+x^2+y^2:x:y:nx:ny
# neumann
c=2*(x*nx+y*ny):x:y:nx:ny
# mu: diffusion term (laplacian) (1)
mu=1
In the Neumann case, we do have the issue mentioned above regarding the definition of the boundary and in particular the unit exterior normal to the boundary. To remedy this problem, we compute the error with respect to the solution defined on \(\Omega_h\) which is a polygon and not a circle. Hence the use of (nx,ny) which provide the normal components at the current point on the boundary.
|
We are now ready to run the application.
$ cd /usr/local/share/feelpp/testcases/quickstart/laplacian/circle
$ mpirun -np 4 feelpp_qs_laplacian_2d --config-file circle-neumann.cfg| The error norms reached machine precision, the solution is in the finite element space. It is due to the fact that (i) we used \(P^2\) finite elements, (ii) the solution is itself quadratic and (iii) we used Dirichlet boundary conditions which requires only point-wise evaluation of the solution on the curved boundary. The fact that the boundary is curved do not affect the numerical solution. It won’t be the case in the presence of Neumann and Robin conditions. |
This gives us some data such as solution of our problem or the mesh used in the application which we can visualize using Paraview. The solution is the same as in [fig:circle-1].
4.2. The case \(\partial \Omega=\partial \Omega_D \cup \partial \Omega_N \cup \partial \Omega_R\)
We finish with all boundary condition types on \(\partial \Omega\) which means that \(\partial \Omega_D\) \(\partial \Omega_R\) and \(\partial \Omega_N\) are non-empty sets. To this end, we mark all boundary sections (1,2) as Dirichlet, (3) as Neumann and (5) as Robin as shown in the Gmsh geo file below
circle-all.geoPhysical Line("Dirichlet") = {1, 2};
Physical Line("Neumann") = {3};
Physical Line("Robin") = {5};
Physical Surface(8) = {7};The next step is to provide the proper conditions or expressions to Feel++ application feelpp_qs_laplacian_2d. They are described in the .cfg file.
circle-all.cfg[functions]
# dirichlet
g=x^2+y^2:x:y
# right hand side
f=-4
# robin
a=1
b=2*(x*nx+y*ny)+x^2+y^2:x:y:nx:ny
# neumann
c=2*(x*nx+y*ny):x:y:nx:ny
# mu: diffusion term (laplacian) (1)
mu=1
In the Neumann case, we do have the issue mentioned above regarding the definition of the boundary and in particular the unit exterior normal to the boundary. To remedy this problem, we compute the error with respect to the solution defined on \(\Omega_h\) which is a polygon and not a circle. Hence the use of (nx,ny) which provide the normal components at the current point on the boundary.
|
We are now ready to run the application.
$ cd /usr/local/share/feelpp/testcases/quickstart/laplacian/circle
$ mpirun -np 4 feelpp_qs_laplacian_2d --config-file circle-all.cfg| The error norms reached machine precision, the solution is in the finite element space. It is due to the fact that (i) we used \(P^2\) finite elements, (ii) the solution is itself quadratic and (iii) we used Dirichlet boundary conditions which requires only point-wise evaluation of the solution on the curved boundary. The fact that the boundary is curved do not affect the numerical solution. It won’t be the case in the presence of Neumann and Robin conditions. |
This gives us some data such as solution of our problem or the mesh used in the application which we can visualize using Paraview. The solution is the same as in [fig:circle-1].
4.3. feelpp2d and feelpp3d
This testcase solves the Laplacian problem in \(\Omega\) an quadrangle or hexadra containing the letters of Feel++